PyBioMed
A python package for generating various molecular representations for chemicals, proteins, DNAs and their interactions
PyBioMed
Main Page: http://projects.scbdd.com/pybiomed.html
GitHub: https://github.com/gadsbyfly/PyBioMed
Installation: https://github.com/gadsbyfly/PyBioMed
Documentation: http://projects.scbdd.com/pybiomed/index.html
Current release: Version 1.0 (2017-9-1)
Introduction
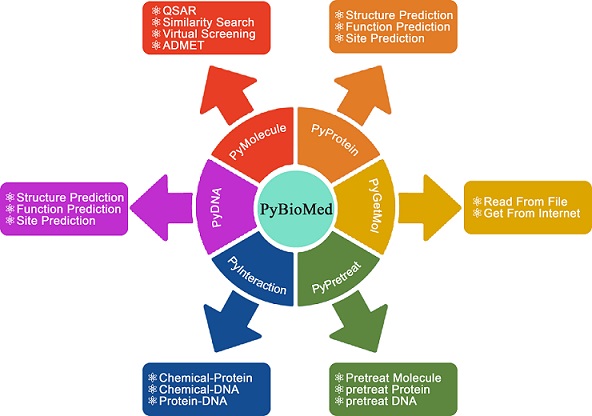
To develop a powerful model for prediction tasks, one of the most important things to consider is how to effectively represent the molecules under investigation such as small molecules, proteins, DNA and even complex interactions, by a descriptor. With PyBioMed you can analyze and represent various complex molecular data under investigation. We hope that the packages will be helpful when exploring questions concerning structures, functions and interactions of various molecular data in the context of systems biology.PyBioMed is a feature-rich python package used for the characterization of various complex biological molecules and interaction samples, such as chemicals,proteins,DNA and their interactions. PyBioMed calculates nine types of features including chemical descriptors or molecular fingerprints, structural and physicochemical features of proteins and peptides from amino acid sequence, composition and physicochemical features of DNA from their primary sequences, chemical-chemical interaction features, chemical-protein interaction features, chemical-DNA interaction features, protein-protein interaction features, protein-DNA interaction features, and DNA-DNA interaction features. PyBioMed can also pretreating molecule structures, protein sequences and DNA sequence. In order to be convenient to users, PyBioMed provides the module to get molecule structures, protein sequence and DNA sequence from Internet.
Features
- Tools for pretreating molecules, proteins sequence and DNA sequence
- Calculating chemical descriptors or molecular fingerprints from molecules’ structures
- Calculating structural and physicochemical features of proteins and peptides from amino acid sequence
- Calculating composition and physicochemical features of DNA from their primary sequences
- Calculating interaction features including chemical-chemical interaction features, chemical-protein interaction features, chemical-DNA interaction features, protein-protein interaction features, protein-DNA interaction features and DNA-DNA interaction features
- Getting molecular structures, protein sequence and DNA sequence from Internet through the molecular ID, protein ID and DNA ID
Function
PyBioMed allows users to compute the characterization of various complex biological molecules and interaction samples, such as chemicals,proteins,DNA and their interactions. (for details see the table).
Developed by

Download
- Download Version 1.0
- Download Documentation
- Download The introduction of molecular descriptors
- Download The introduction of protein descriptors
- Download The introduction of DNA descriptors
- Download The introduction of interaction descriptors
Overview
Related links
Software
Github (latest development): >>Click here
Documentation
The introduction of descriptors
The introduction of molecular descriptors
The introduction of protein descriptors
The introduction of DNA descriptors
The introduction of interaction descriptors