ChemDes
An integrated web-based platform for molecular descriptor and fingerprint computation.
ChemDes
Main Page: http://www.scbdd.com/chemdes
Current release: Version 3.0 (2014-10-20)
Introduction
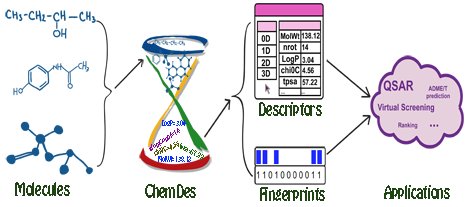
ChemDes is a free web-based platform for the calculation of molecular descriptors and fingerprints, which provides more than 3,679 molecular descriptors that are divided into 61 logical blocks.In addition, it provides 59 types of molecular fingerprint systems for drug molecules, including topological fingerprints, electro-topological state (E-state) fingerprints, MACCS keys, FP4 keys, atom pairs fingerprints, topological torsion fingerprints and Morgan/circular fingerprints, et al.
Features
- ChemDes is freely available to the public and requires no programming skills.
- ChemDes has integrated various molecular descriptors and fingerprints from the toolkits written in different programming languages (C++, Python and Java...).
- ChemDes integrates MOPAC software and incorporates three useful tools (ChemCONV, ChemMOP and ChemFPS).
- ChemDes possesses advantages of cross-platform and interoperability. Users can access this platform via almost all the operation system types (Microsoft windows, Linux, Mac OS, Android) and client types (PC clients, mobile clients); The calculating results and input/output files from ChemDes can be directly used in other calculations or studies.
Functionalities
Molecular descriptors
ChemDes allows users to compute 3679 molecular descriptors from several open source packages (for details see Molecular descriptors library).
- Chemopy Descriptors 1135
- CDK Descriptors 275
- RDKit Descriptors 196
- Pybel Descriptors 24
- BlueDesc Descriptors 174
- PaDEL Descriptors 1875
Molecular descriptors
ChemDes allows users to compute 59 types of molecular fingerprints (for details see Molecular fingerprints library ).
- Molecular Fingerprints 59
Useful tools
ChemDes allows users to compute 59 types of molecular fingerprints (for details see Molecular fingerprints library ).
Developed by

Download
Overview
Publications
- Jie Dong, Dong-Sheng Cao, Hong-Yu Miao, Shao Liu, Bai-Chuan Deng, Yong-Huan Yun, Ning-Ning Wang, Ai-Ping Lu, Wen-Bin Zeng, Alex Chen. ChemDes: an integrated web-based platform for molecular descriptor and fingerprint computation. Journal of Cheminformatics 2015, 7:60
Latest updates
- Version 1.0 (2013-10-20). The ChemDes Version 1.0 was released
- Version 1.1 (2013-11-20). Added module Molecular Fingerprints.Added the following fingerprints(topological fingerprints, electro-topological state (E-state) fingerprints, MACCS keys, FP4 keys, atom pairs fingerprints, topological torsion fingerprints and Morgan/circular fingerprints.)
- Version 1.1.1(2013-11-27).Fixed bugs.optimized the generating process of the csv file.
- Version 1.2 (2013-12-05). Added tool:ChemCONV.Users can use this to convert the format of molecule and prepare the inputformats
- Version 1.3 (2014-02-07). Added tool:ChemMOP.It is developed based on PyBel and MOPAC2012 to help studying of molecular structures,
- Version 1.4 (2014-03-16). Added tool:ChemFPS.It is a free online-tool developed for Calculating Molecular Similarity based on molecular fingerprints by using measures such as 'Tanimoto'.
- Version 1.5 (2014-07-11). Added new descriptors from CDK.
- Version 1.6 (2014-08-18). Added new descriptors from RDKit.
- Version 1.7 (2014-10-06). Added new descriptors from Pybel.
- Version 1.8 (2014-11-11). Added new descriptors from BlueDesc.
- Version 1.9 (2014-11-17). Added new descriptors from PaDEL.
- Version 2.0(2014-11-23). Update the tool ChemFPS. Added the batch computing function.
- Version 3.0(2014-12-01). Update the module Molecular Fingerprints . Added the following fingerprints('FP2 fingerprints', 'FP3 fingerprints', 'RDKit fingerprints', 'Pattern fingerprints', 'Layered fingerprints', 'Pubchem fingerprints', 'CDK fingerprints', 'CDK extended fingerprints' 'Klekota-Roth fingerprints', 'Klekota-Roth fingerprint count', 'CDK graph only fingerprints', 'Substructure fingerprints', 'Substructure fingerprint count', '2D atom pairs', '2D atom pairs count', 'Hybridization fingerprints'.).
- Version 3.0(2016-11-09). Fixed bugs and updated MOPAC to MOPAC2016.